Rafe Helwer1,2,3 , and J. Michael Charette1,2,3§
, and J. Michael Charette1,2,3§
1Department of Chemistry, Brandon University, Brandon, Manitoba, Canada.
2Children’s Hospital Research Institute of Manitoba, Winnipeg, Manitoba, Canada.
3CancerCare Manitoba Research Institute, Winnipeg, Manitoba, Canada.
§Correspondence to: J. Michael Charette (charettem@brandonu.ca)
Abstract
Description
Methods
Extended Data
- Description: Extended figure 1: The N-terminal region of the yeast and human Utp25 is disordered. (Top) PONDR prediction of an N-terminal IDR (aa 1 to ~160) in the yeast Utp25. (Bottom) PONDR prediction of an N-terminal IDR (aa 1 to ~185) in the human UTP25. Resource Type: Image. DOI: 10.22002/D1.20292
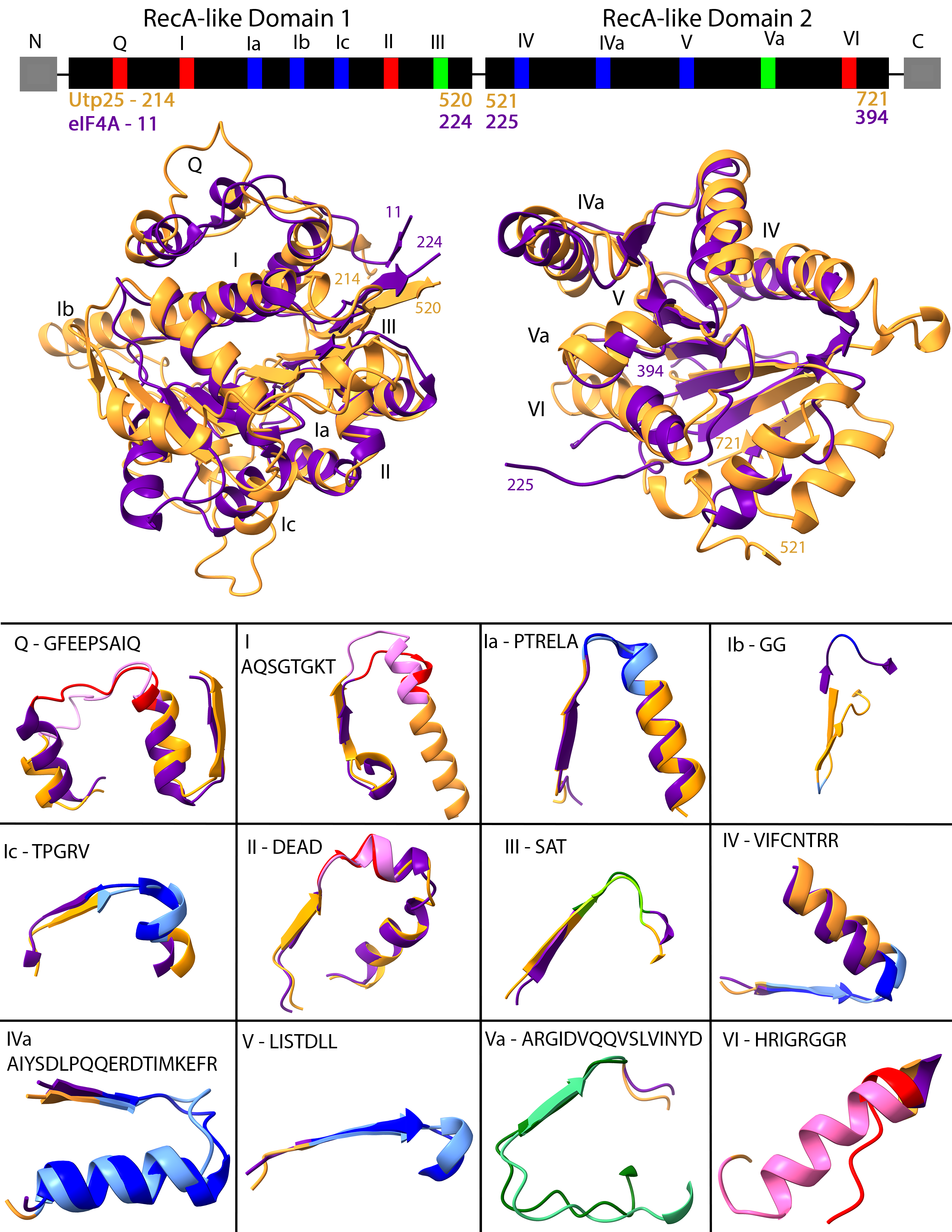
- Description: Extended figure 2: The high pLDDT domains 1 and 2 of the AlphaFold yeast Utp25 align well to eIF4A. Structural alignment of the individual RecA-like domains 1 and 2 of the AlphaFold yeast Utp25 (P40498) and yeast eIF4A (1FUU; (Caruthers et al. 2000)) crystal structure. The AlphaFold yeast Utp25 is coloured based on pLDDT score, from very low confidence in red to very high confidence in blue (key in bottom right) whereas the yeast eIF4A crystal structure is shown in brown. In domain 1, six regions in the AlphaFold yeast Utp25 are coloured cyan (70 < pLDDT < 90; confident) as opposed to blue (pLDDT > 90, very high confidence). Two of these areas overlap with helicase motifs Q and Ib. Similarly, there are 7 confident areas in domain 2 (cyan), with one overlapping helicase motif VI. The remainder of the helicase motifs are in high confidence areas. The location of the helicase motifs is indicated along with the coordinates of the regions used in the structural alignments of isolated domains 1 and 2. Resource Type: Image. DOI: 10.22002/D1.20293
- Description: Extended figure 3: The yeast and human AlphaFold Utp25 structures are conserved. ChimeraX file of the aligned yeast (P40498) and human (Q68CQ4) AlphaFold Utp25 structures shows that they are clearly superimposable and thus conserved in structure. Resource Type: InteractiveResource. DOI: 10.22002/D1.20294
- Description: Extended figure 4: Dali search using the yeast and human AlphaFold Utp25 structures robustly identifies known DEAD-box helicases. Resource Type: Dataset. DOI: 10.22002/D1.20295
- Description: Extended figure 5: Alignment of the full-length Utp25 and eIF4A structures. ChimeraX file of the aligned full-length (containing RecA domains 1 and 2) AlphaFold yeast Utp25 (P40498) and yeast eIF4A (1FUU; (Caruthers et al. 2000)) crystal structure. Note the structural similarity of domains 1 of Utp25 and of eIF4A and similarly of domains 2 of both proteins. Structural overlap is only possible for one or the other of the two RecA-like domains but not for both domains simultaneously. Resource Type: InteractiveResource. DOI: 10.22002/D1.20296
- Description: Extended figure 6: Alignment of the RecA-like domains 1 of Utp25 and of eIF4A. ChimeraX file of the aligned domains 1 of the AlphaFold yeast Utp25 (P40498) and yeast eIF4A (1FUU; (Caruthers et al. 2000)) crystal structure. Resource Type: InteractiveResource. DOI: 10.22002/D1.20297
- Description: Extended figure 7: Alignment of the RecA-like domains 2 of Utp25 and of eIF4A. ChimeraX file of the aligned domains 2 of the AlphaFold yeast Utp25 (P40498) and yeast eIF4A (1FUU; (Caruthers et al. 2000)) crystal structure. Resource Type: InteractiveResource. DOI: 10.22002/D1.20298
Acknowledgements
Funding
Author Contributions
- Rafe Helwer: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing - original draft
- J. Michael Charette: Conceptualization, Funding acquisition, Formal analysis, Supervision, Writing - review & editing
Reviewed By
Anonymous, Joel Sussman
History
- Received: 7/8/2022
- Revision Received: 8/19/2022
- Accepted: 9/19/2022
- Published Online: 9/22/2022
- Indexed: 10/6/2022
Copyright
© 2022 by the authors. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International (CC BY 4.0) License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation
PubMed Central: 9539457
PubMed: 36212518
microPublication Biology:ISSN: 2578-9430

